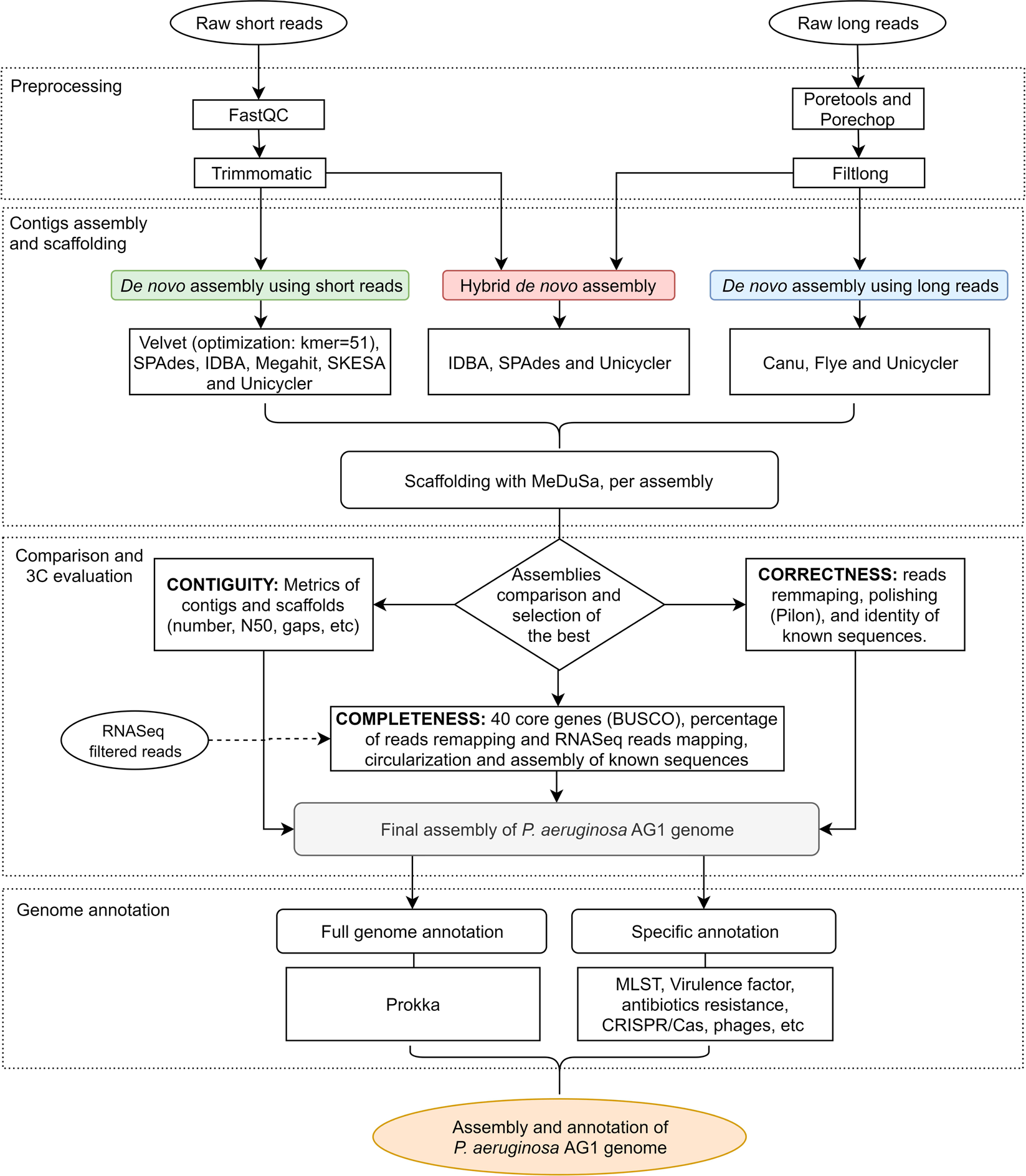
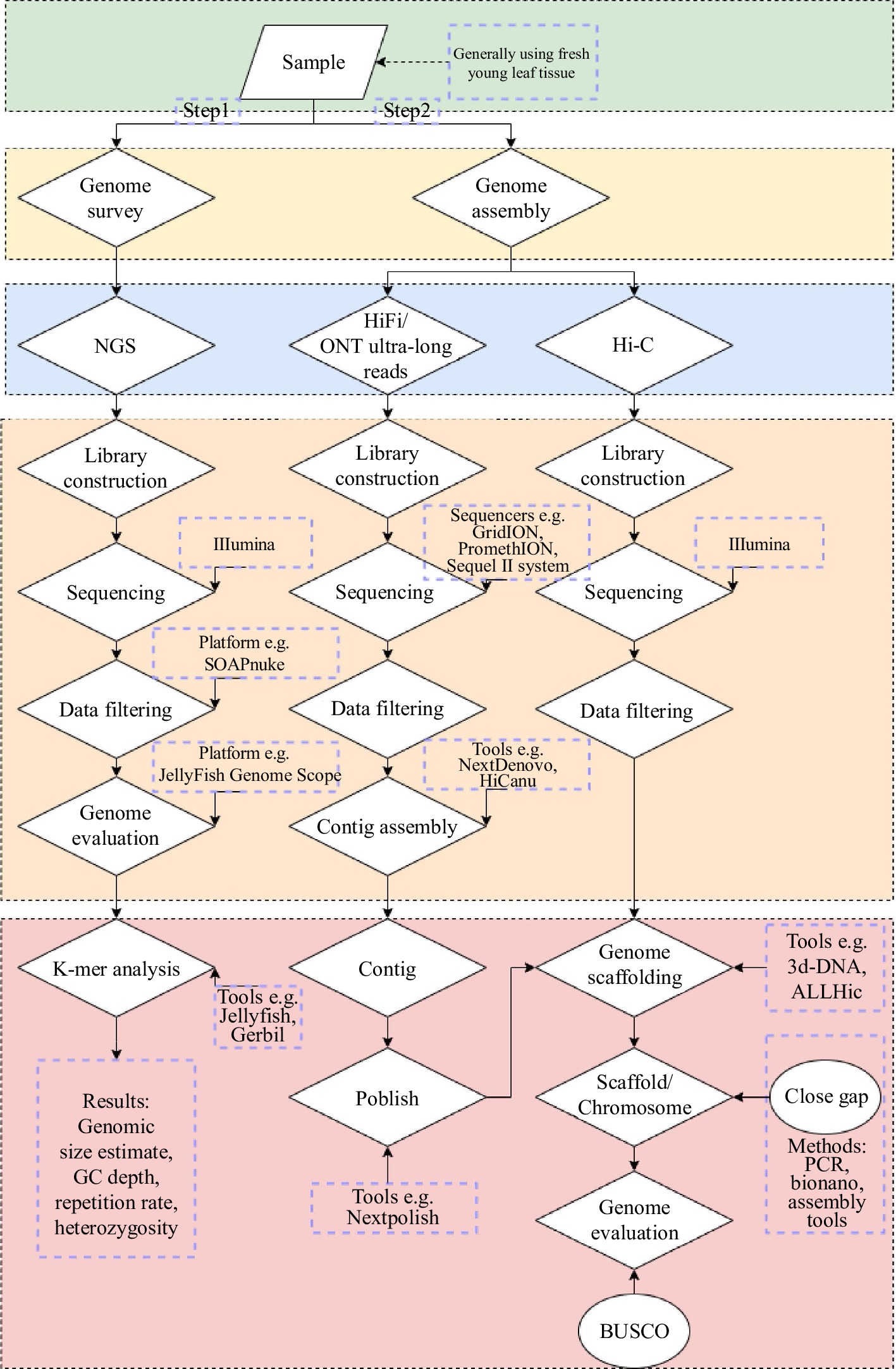
GAPPadder: a sensitive approach for closing gaps on draft genomes with short sequence reads | BMC Genomics | Full Text
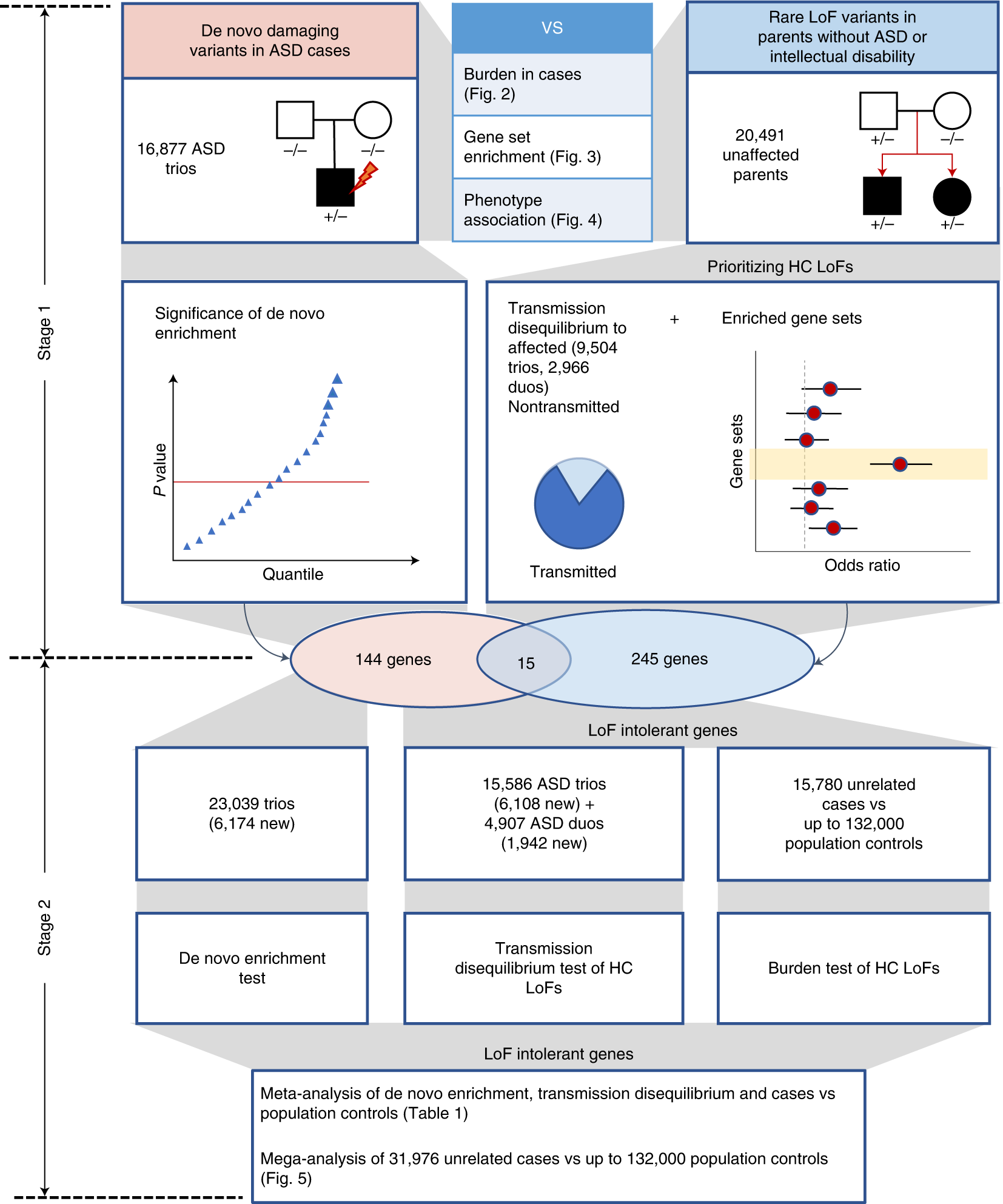
Integrating de novo and inherited variants in 42,607 autism cases identifies mutations in new moderate-risk genes | Nature Genetics

Expanding the space of protein geometries by computational design of de novo fold families | Science
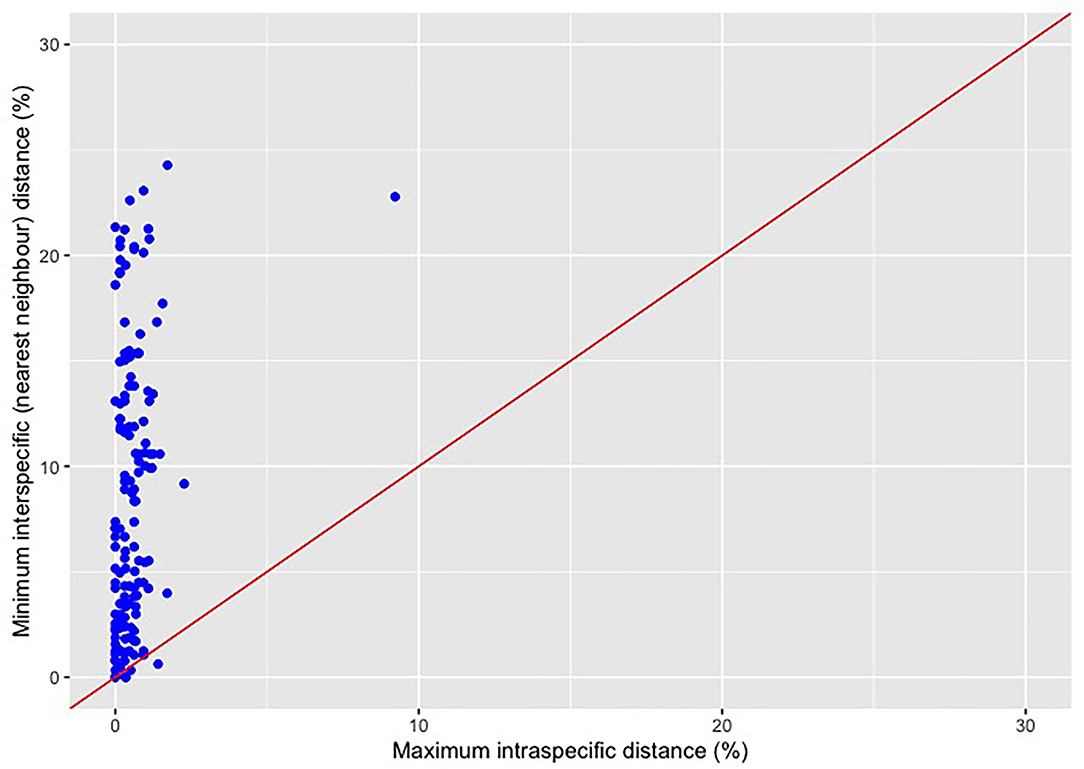
Frontiers | Lack of Statistical Rigor in DNA Barcoding Likely Invalidates the Presence of a True Species' Barcode Gap
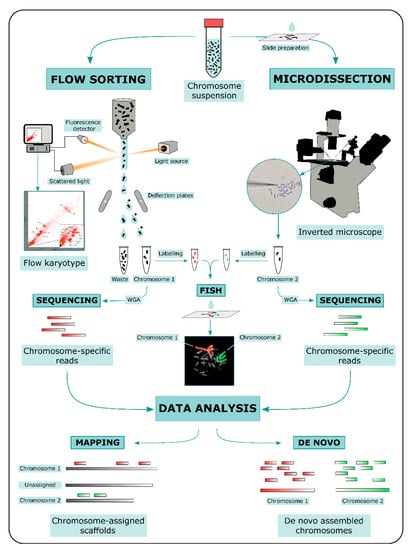
Genes | Free Full-Text | Bridging the Gap between Vertebrate Cytogenetics and Genomics with Single-Chromosome Sequencing (ChromSeq)

Development of a conceptual model and patient-reported outcome measures for assessing symptoms and functioning in patients with heart failure | Quality of Life Research
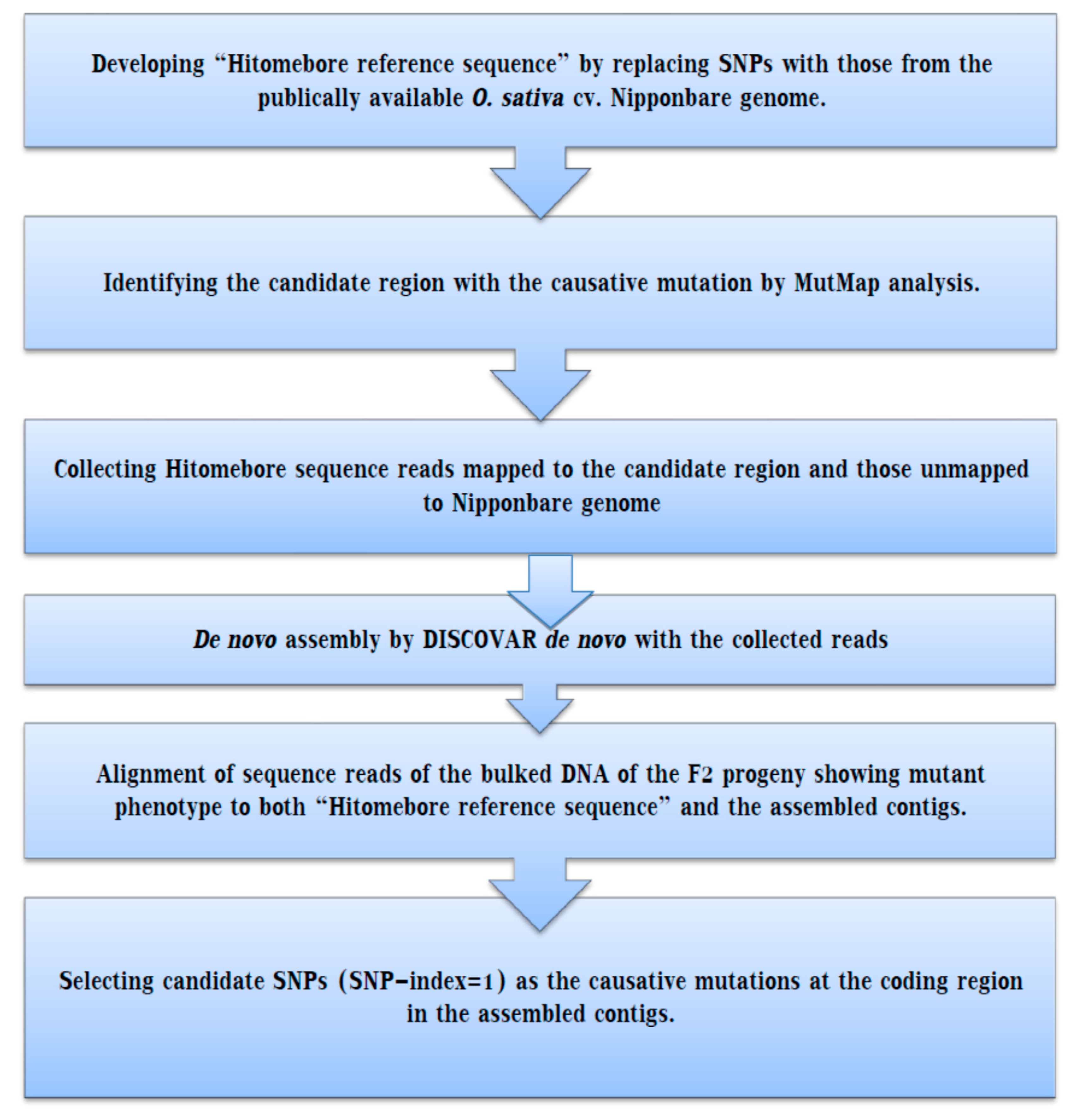
IJMS | Free Full-Text | Whole Genome Resequencing from Bulked Populations as a Rapid QTL and Gene Identification Method in Rice

High-throughput pipeline for the de novo viral genome assembly and identification of minority variants from Next-Generation Sequencing of residual diagnostic samples | bioRxiv

De novo and recurrent metastatic breast cancer – A systematic review of population-level changes in survival since 1995 - eClinicalMedicine

Application of de Novo Sequencing to Large-Scale Complex Proteomics Data Sets | Journal of Proteome Research

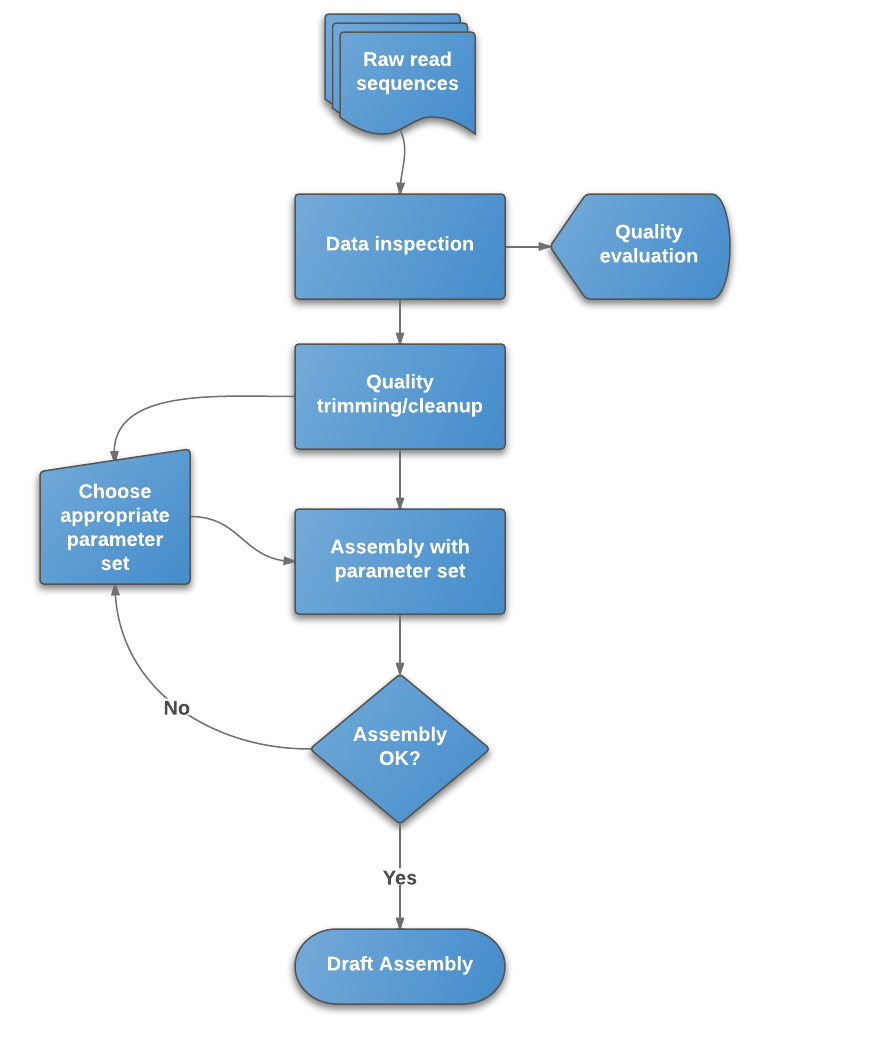
Workflow of a typical de novo whole-genome sequencing project. Black... | Download Scientific Diagram
Selecting Superior De Novo Transcriptome Assemblies: Lessons Learned by Leveraging the Best Plant Genome | PLOS ONE
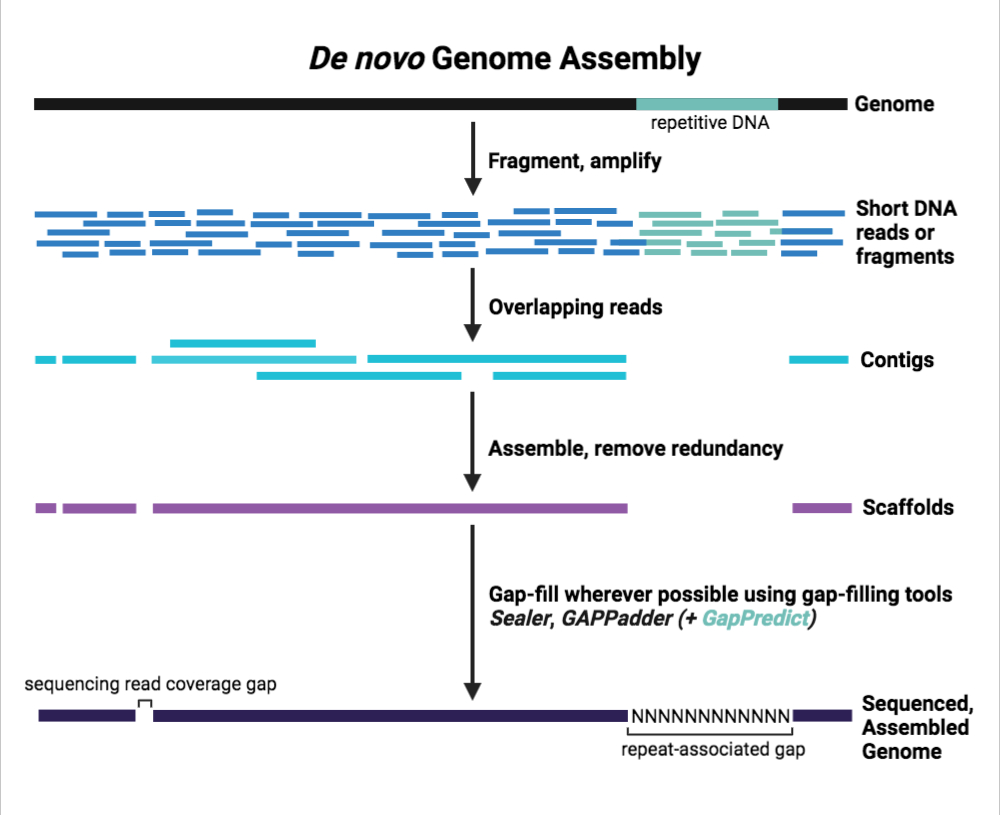
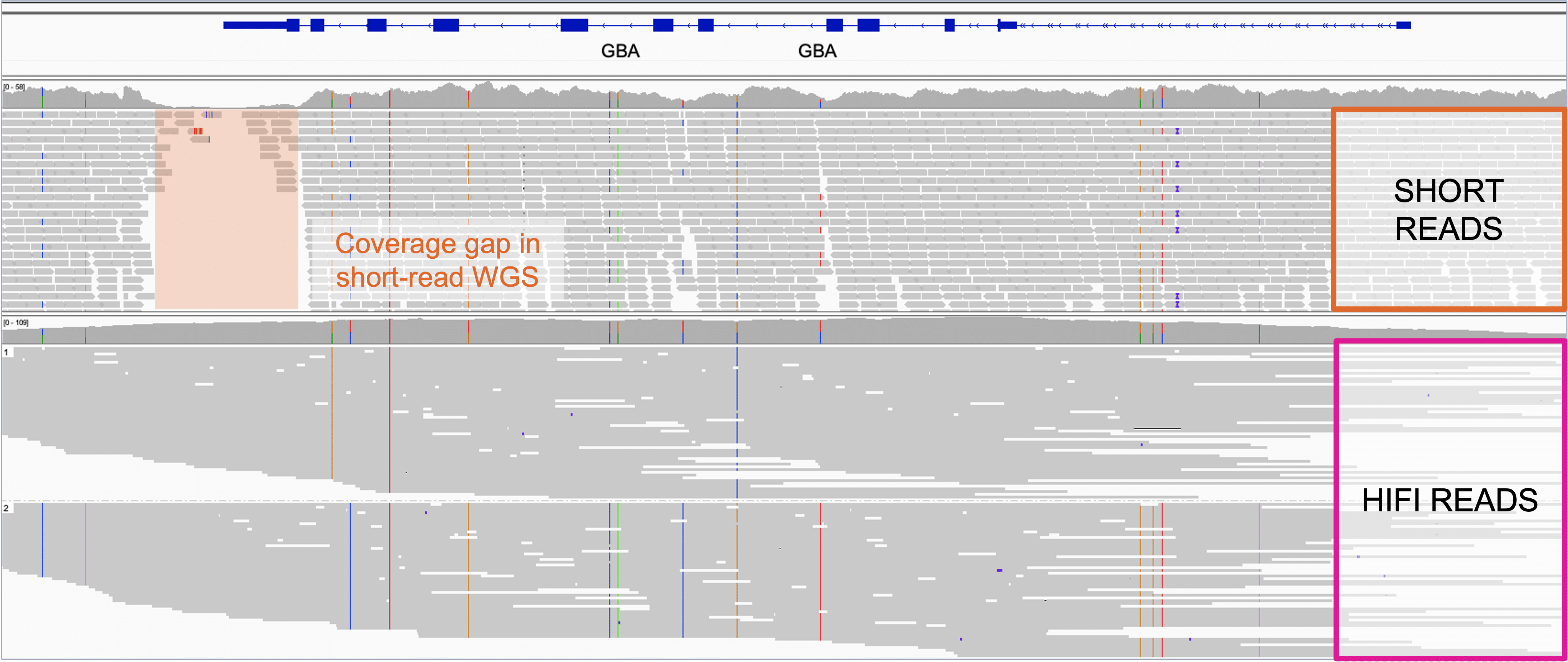
Filling in the gaps: GapPredict can complement repertoire of tools used to resolve missing DNA sequences in genome assemblies | Genome Sciences Centre

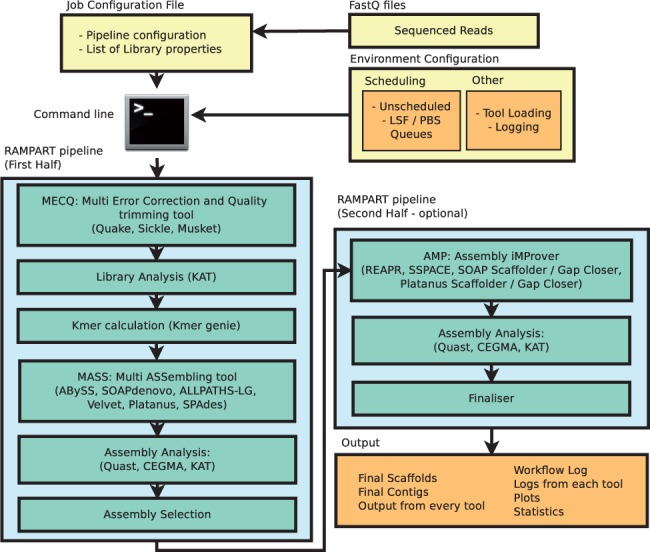
De novo assembly. High-level diagram of long-read assembly pipeline.... | Download Scientific Diagram

A pipeline for completing bacterial genomes using in silicoand wet lab approaches | BMC Genomics | Full Text

Integrated gene analyses of de novo variants from 46,612 trios with autism and developmental disorders | PNAS

The impact of noise and missing fragmentation cleavages on de novo peptide identification algorithms - ScienceDirect